Mécanismes de la traduction
Évolution du démarrage de la traduction
L'étude des processus fondamentaux, comme la biosynthèse des protéines, permet de mieux comprendre l'évolution. Ainsi, l'analyse des séquences de l’ARN ribosomal de la petite sous-unité a conduit à définir un arbre du vivant organisé en trois domaines, bactéries, eucaryotes et archées. Au sein de cet arbre les archées sont proches des eucaryotes et apparaissent comme une pièce centrale pour comprendre l'évolution des organismes vivants. Les efforts pour mieux comprendre la diversité des archées et de leurs mécanismes moléculaires apporteront des informations importantes.
Chez les archées, les ARNm ne sont pas modifiés après leur transcription, comme chez les bactéries. Selon le phylum d’archée, les ARNm portent des séquences Shine-Dalgarno ou ont des régions 5' non traduites très courtes (sans leader). Cependant, d'autres caractéristiques de l'appareil de traduction rapprochent les archées des eucaryotes. En particulier, le ribosome archéen et les facteurs de traduction sont de type eucaryote.
Notre équipe s'intéresse principalement aux complexes ribosomaux d'initiation de la traduction. Nous étudions les organismes archéens et nous nous intéressons également aux bactéries et aux eucaryotes pour mieux comprendre comment les mécanismes d'initiation de la traduction ont évolué. Nous utilisons la cristallographie aux rayons X, le SAXS et la cryo-EM pour réaliser des études structurales et nous avons également développé des techniques biochimiques pour les études fonctionnelles.
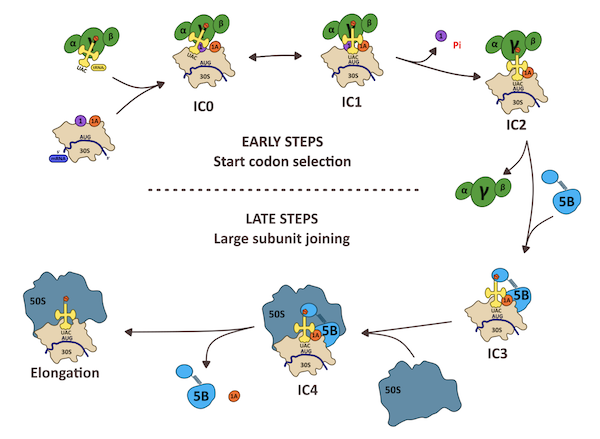
Étapes du démarrage de la traduction chez les archées
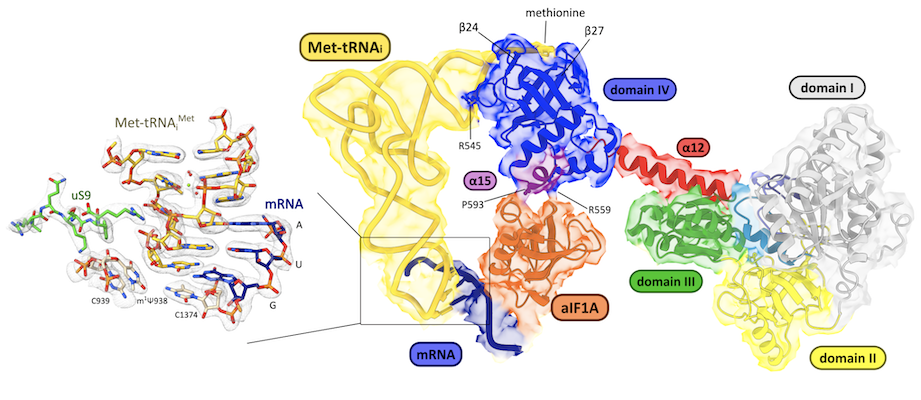
Structure cryo-EM du complexe IC3 à 2.7 Å montrant l’interaction des facteurs de démarrage avec la petite sous-unité du ribosome (Kazan R, et. al., Nucleic Acids Research 2022)
Ingénierie des protéines
L'incorporation spécifique d'un acide aminé beta dans un polypeptide introduirait une flexibilité sans précédent dans la chaîne principale, élargissant ainsi les possibilités géométriques de repliement des protéines. Il a récemment été démontré, à l'aide d'un système de traduction in vitro sans cellules, que le ribosome était capable d'incorporer efficacement un acide aminé beta dans un polypeptide. Ainsi, pour permettre la synthèse in vivo de protéines contenant des acides-aminés beta il reste à concevoir des aminoacyl-ARNt synthétases capables d'estérifier efficacement ces acides aminés beta sur un ARNt spécifique. Nous travaillons sur ce problème en collaboration avec Philippe Marlière (Heurisko INC, USA) et Valérie Pezo (ISSB, Evry) en utilisant la méthionyl-tRNA synthétase (MetRS) de E. coli. Les études biochimiques et structurales de la liaison de la méthionine à la MetRS donnent des indices intéressants pour renforcer l’activité catalytique de l’enzyme vis-à-vis des acides aminés beta, avec notamment une structure cristallographique à 1,4 Å de résolution pour un complexe MetRS:beta-Met.
eIF2 dans les maladies cardiaques
Le stress du réticulum endoplasmique (RE) est apparu comme un mécanisme important impliqué dans la pathogenèse des maladies cardiovasculaires. eIF2 est un facteur impliqué dans le démarrage de la traduction, mais il est aussi un effecteur clé de la réponse au stress du RE. La perturbation de la fonction du RE entraîne une accumulation de protéines, ce qui déclenche la « réponse aux protéines non repliées » (UPR). Au cours de cette réponse, eIF2 est phosphorylé sur sa sous-unité alpha par la kinase PERK. Cela déclenche une série d'événements de transduction pour réduire l'accumulation de protéines et le stress du RE. Une induction légère à modérée de cette voie favorise une réponse restauratrice alors qu'une réponse sévère ou chronique au stress du RE favorise l'apoptose et contribue au développement de la maladie cardiaque.
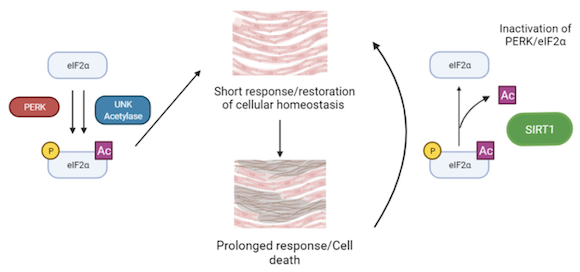
Une découverte récente de notre collaborateur, C. Lemaire (INSERM UMR-S1180), a montré que cette voie était contrôlée non seulement par la phosphorylation d'eIF2, mais aussi par son acétylation. De plus, la désacétylation d'eIF2 par la protéine sirtuin 1 (SIRT1), protège les cardiomyocytes de l'apoptose induite par le stress ER. La voie SIRT1-eIF2 pourrait être utilisée en thérapie médicamenteuse, car les taux d'activité de SIRT1 peuvent être modifiés par des composés activateurs de sirtuines (STACs). Notre objectif est de comprendre les bases moléculaires de l'interaction de SIRT1 avec eIF2α, ainsi que l'interaction entre l'acétylation et la phosphorylation d'eIF2α.
Collaborations :
- SOLEIL Synchrotron (TREMTI project supported by ANR)
- Anh Tuân Phan (NTU, Singapore)
- Philippe Marlière (Heurisko INC, USA) and Valérie Pezo (ISSB, Evry)
- Béatrice Clouet D’Orval (CBI, Toulouse)
- Christophe Lemaire (INSERM UMR-S1180)
Sélection de publications :
- Kazan R, Bourgeois G, Lazennec-Schurdevin C, Larquet E, Mechulam Y, Coureux PD, Schmitt E. Role of aIF5B in archaeal translation initiation. Nucleic Acids Res. 2022, 50, 6532–6548, doi.org/10.1093/nar/gkac490.
- Nigro G, Bourcier S, Lazennec-Schurdevin C, Schmitt E, Marlière P, Mechulam Y. Use of β3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase. J Struct Biol. 2020, 209, 107435, doi.org/10.1016/j.jsb.2019.107435.
- Coureux PD, Lazennec-Schurdevin C, Bourcier S, Mechulam Y, Schmitt E. Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation. Commun Biol. 2020, 3, 58, doi.org/10.1038/s42003-020-0780-0.
- Schmitt, E, Coureux, P-D, Kazan, R., Bourgeois, G., Lazennec-Schurdevin, C., Mechulam, Y. Recent advances in archaeal translation initiation. Frontiers in Microbiology, section Biology of Archaea. Front Microbiol. 2020 Sep 18;11:584152, doi.org/10.3389/fmicb.2020.584152.


