Nanoimaging for cell dynamics and quantitative biology
Our web site has moved to this address: https://lob.ip-paris.fr
Permanent staff
Antigoni Alexandrou (senior research scientist - CNRS)
Cédric Bouzigues (assistant professor - Polytechnique)
Nicolas Olivier (research scientist - CNRS)
Rivo Ramodiharilafy (assistant engineer - Polytechnique)
Position proposals |
|
Ph. D. Thesis/Internship proposal: In vivo detection of reactive oxygen species using multifunctional luminescent nanoparticles Applications from highly talented and motivated candidates are encouraged. |
Research topics
We develop non-blinking single-molecule labels and sensors based on lanthanide-ion doped nanoparticles and use them for studying toxin-cell interaction and toxin receptor confinement in the cell membrane, and hydrogen peroxide production in signaling processes, respectively. We also develop and use microfluidic devices to produce asymmetric stimulation of cells and induce controlled deoxygenation of red blood cells in microfluidic droplets.
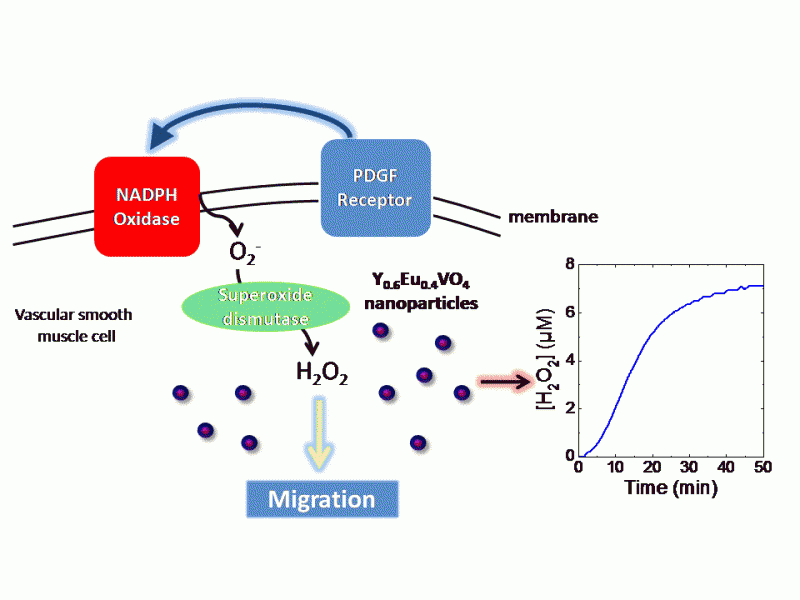
These nanoparticles are particularly attractive for single-molecule tracking applications (Nano Lett. 2004, Phys. Rev. Lett. 2009): they are synthesized directly in water, present high photostability and no blinking, narrow emission linewidths independent of nanoparticle size, and long excited state lifIntracellular H2O2 detection using Eu-doped nanoparticlesetimes which are useful for retarded detection schemes and FRET applications (J. Phys. Chem. B 2006). We furthermore discovered that Eu-doped nanoparticles are oxydant probes and can be used for quantitative time-resolved intracellular H2O2 detection with spatial resolution (Nat. Nanotech. 2009, Chem. Biol. 2014).
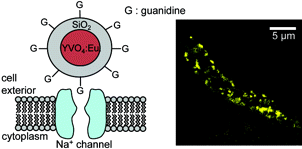
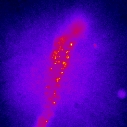
We demonstrated that these nanoparticles are detectable individually and that their size can be accurately determined from their luminosity (Appl. Phys. Lett. 2006). We have shown that nanoparticles functionalized with guanidine groups specifically target sodium channels and can be imaged individually on living cardiomyocytes (Nano Lett. 2004). We then implemented a more general nanoparticle-protein coupling scheme and counted the number of proteins coupled to individual nanoparticles (J. Am. Chem. Soc. 2007).
Nanoparticles labeling sodium channels on the membrane of a frog cardiomyocyte
We now use peptidic toxins (family of oligomerizing and pore forming toxins) labeled by Eu-doped nanoparticles to investigate their interaction with cells: binding to receptors on the cell membrane and receptor motion inside membrane microdomains. Novel single-molecule trajectory analysis techniques based on Bayesian inferences can extract the force field felt by the receptor (Phys. Rev. Lett. 2009, Biophys. J. 2010, 2012). These toxins can thus also be viewed as tools for investigating the membrane organization.
Low concentrations of reactive oxygen species and, in particular hydrogen peroxide (H2O2), mediate numerous signaling processes in the cell. We measured the differences in the timing of intracellular H2O2 production triggered by different signals using Eu-doped nanoprobes (Nat. Nanotech 2009). We are now using this novel sensor to investigate the role of receptor transactivation and asymmetric stimulation in migration and contraction processes (Chem. Biol. 2014).
Dynamics of sickle red blood cells in microfluidic channels
Microfluidics offers the opportunity of generating controlled model environments reproducing physiological conditions while using minimal amounts of sample solutions. In particular, microfluidic channels are ideal for the study of blood cells in conditions approaching those of the blood flow.
We investigate the dynamics of red blood cells in the case of sickle cell disease, a genetic disease involving a single point mutation of the oxygen-carrying protein, hemoglobin. This mutation induces hemoglobin polymerization upon de-oxygenation which leads to a decrease in the deformability of red blood cells. This in turn leads to painful, vaso-occlusion episodes, organ damage and a shortened lifespan. We have implemented an innovating microfluidic platform that can generate cycles of oxygenation-deoxygenation close to the physiological ones to study this disease at the single cell level. Using polarization microscopy, we detect the hemoglobin polymerization by the birefringence it induces and study the behavior of numerous single red blood cells and the effect of important parameters and inhibitor molecules. This study should bring insight into the molecular mechanisms of the disease and help develop new therapeutic approaches.
Former members :
Rachid Rezgui (Ph. D. student, 2010-2013)
Paul Abbyad (Post-doc, 2008-2012)
Markus Schöffel (Ph. D. student, 2009-2012)
Silvan Türkcan (Ph. D. student, 2007-2010 and post-doc 2011-2012)
Thanh-Liêm Nguyên (Ph.D. student, 2006-2009)
Didier Casanova (Ph. D. student, 2004-2007)
Egidius Auksorius (Marie-Curie exchange Ph. D. student, 2004)
Undergraduate students : G. Demangeon (2006), T. Amirtha (2005), T. Kourkoutsaki
Collaborations :
T. Gacoin, J.-P. Boilot, Lab. of condensed matter physics (Ecole Polytechnique, Palaiseau)
J.-M. Allain, Lab. de Mécanique des Solides (Ecole Polytechnique, Palaiseau)
M. Popoff, Unité Bactéries Anaérobies et Toxines (Institut Pasteur, Paris)
J.-B. Masson, M. Vergassola, Physics of Biological systems (Institut Pasteur, Paris)
P.-L. Tharaux, INSERM U970, Centre de Recherche Cardiovasculaire de Paris-PARCC