Translation mechanisms
Evolution of translation initiation
Studies of fundamental processes, such as protein biosynthesis, provide a better understanding of evolution. Thus, the analysis of ribosomal RNA sequences of the small ribosomal subunit has led to a tree of life organized in three domains, bacteria, eukaryotes and archaea. Within this tree, archaea are close to eukaryotes and appear as a central piece to understand the evolution of living organisms. Efforts to better understand the diversity of archaea and their molecular mechanisms will provide important information
In archaea, mRNAs are not matured after their transcription, as in bacteria. Depending on the archaeal phylum, either mRNAs carrying Shine-Dalgarno sequences or mRNAs having very short 5' untranslated regions (leaderless) are preferentially used. However, other features of the translation apparatus bring archaea closer to eukaryotes. In particular, the archaeal ribosome and translation factors are of the eukaryotic type.
Our team is mainly interested in ribosomal complexes of translation initiation. We study archaeal organisms and are also interested in bacteria and eukaryotes to better understand how the translation initiation mechanisms have evolved. We use X-ray crystallography, SAXS and cryo-EM to perform structural studies and we also developed biochemical strategies for functional studies.
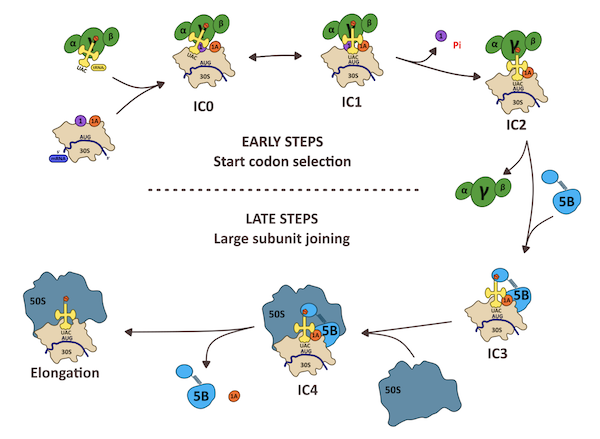
Steps of translation initiation in Archaea
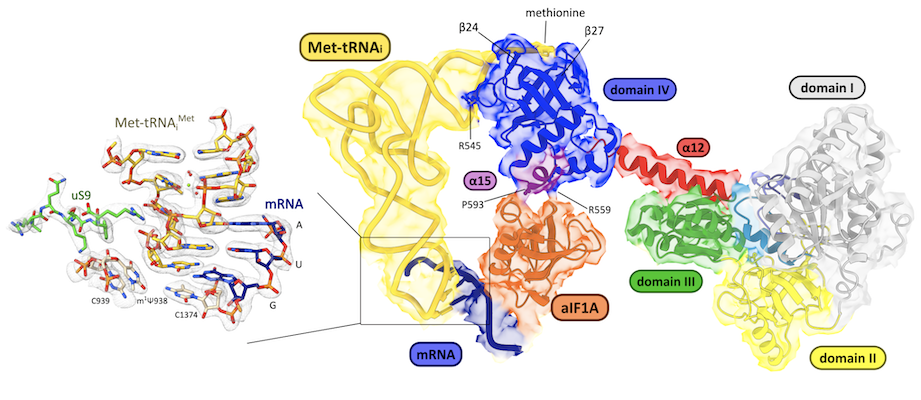
Cryo-EM structure of the IC3 complex at 2.9 Å resolution showing the interaction of the initiation factors with the small ribosomal subunit (Kazan R, et. al., Nucleic Acids Research 2022)
Protein Engineering
Site-specific incorporation of a β amino acid in a polypeptide would introduce flexibility in the main chain, thereby enlarging the geometric possibilities for protein folding. It has recently been demonstrated, using an in vitro cell-free translation system, that the ribosome is able to efficiently incorporate a β amino acid in a polypeptide. To enable in vivo synthesis of proteins containing β residues it is still necessary to design aminoacyl-tRNA synthetases capable of efficiently esterifying β amino acids on a specific tRNA. We work on this problem in collaboration with Philippe Marlière (Heurisko INC, USA) and Valérie Pezo (ISSB, Evry) using E. coli methionyl-tRNA synthetase (MetRS). Biochemical and structural studies of β methionine binding to MetRS give interesting clues for improving the catalytic activity of the enzyme towards β amino acids. In particular, we solved a 1.4 Å resolution X-ray structure of a MetRS:β-Met complex.
eIF2 in cardiac diseases
Endoplasmic reticulum (ER) stress has emerged as an important mechanism involved in the pathogenesis of cardiovascular diseases. eIF2 is a factor involved in translation initiation, however it is also a key effector of the ER stress response. Disruption of ER function leads to protein accumulation, which triggers the Unfolded Protein Response (UPR). During this response, eIF2 is phosphorylated in its alpha subunit by the PERK kinase. This triggers a series of transduction events to reduce protein accumulation and ER stress. It is considered that a mild-to-moderate induction of this pathway favours a restorative autophagy-based response whereas severe or chronic ER stress response promotes apoptosis which contributes to the development of cardiac disease.
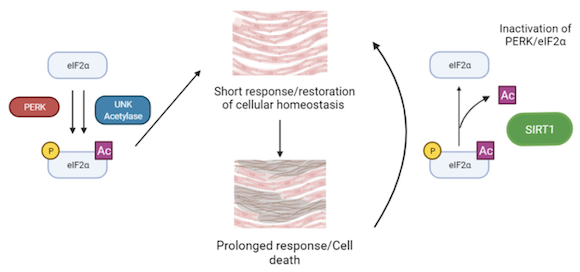
However, a recent discovery by our collaborator, C. Lemaire, showed that this pathway is controlled not only by eIF2 phosphorylation, but also by its acetylation. Moreover, deacetylation of eIF2 by the protein sirtuin 1 (SIRT1), protects cardiomyocytes from ER-stress induced apoptosis, by specifically attenuating activation of the eIF2 phosphorylation pathway and promoting autophagy. Interestingly, phosphorylation and acetylation levels of eIF2 seem to be related as overall increase of eIF2 acetylation is usually paired with an increase of phosphorylated eIF2. This SIRT1-eIF2 pathway has a potential to be used in drug therapy, as SIRT1 activity rates can be modified by Sirtuin Activating Compounds (STACs). Our aim is to understand the molecular basis of the interaction of SIRT1 with eIF2α, as well as the interplay between eIF2α acetylation and phosphorylation.
Main collaborations:
- SOLEIL Synchrotron (TREMTI project supported by ANR)
- Anh Tuân Phan (NTU, Singapore)
- Philippe Marlière (Heurisko INC, USA) and Valérie Pezo (ISSB, Evry)
- Béatrice Clouet D’Orval (CBI, Toulouse)
- Christophe Lemaire (INSERM UMR-S1180)
Selected publications:
- Fedry J, Silva J, Vanevic M, Fronik S, Mechulam Y, Schmitt E, des Georges A, Faller WJ, Förster F. Visualization of translation reorganization upon persistent ribosome collision stress in mammalian cells. Mol Cell. 2024 Mar 21;84(6):1078-1089.e4, doi.org/10.1016/j.molcel.2024.01.015.
- Opuu V, Nigro G, Lazennec-Schurdevin C, Mechulam Y, Schmitt E, Simonson T. Redesigning methionyl-tRNA synthetase for β-methionine activity with adaptive landscape flattening and experiments. Protein Sci. 2023 Sep;32(9):e4738, doi.org/10.1002/pro.4738
- Cardenal Peralta C, Vandroux P, Neumann-Arnold L, Panvert M, Fagart J, Seufert W, Mechulam Y, Schmitt E. Binding of human Cdc123 to eIF2γ. J Struct Biol. 2023 Sep;215(3):108006, doi.org/10.1016/j.jsb.2023.108006.
- Nigro G, Bourcier S, Lazennec-Schurdevin C, Schmitt E, Marlière P, Mechulam Y. Use of β3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase. J Struct Biol. 2020, 209, 107435, doi.org/10.1016/j.jsb.2019.107435.
- Kazan R, Bourgeois G, Lazennec-Schurdevin C, Larquet E, Mechulam Y, Coureux PD, Schmitt E. Role of aIF5B in archaeal translation initiation. Nucleic Acids Res. 2022, 50, 6532–6548, doi.org/10.1093/nar/gkac490.
- Coureux PD, Lazennec-Schurdevin C, Bourcier S, Mechulam Y, Schmitt E. Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation. Commun Biol. 2020, 3, 58, doi.org/10.1038/s42003-020-0780-0.
- Schmitt, E, Coureux, P-D, Kazan, R., Bourgeois, G., Lazennec-Schurdevin, C., Mechulam, Y. Recent advances in archaeal translation initiation. Frontiers in Microbiology, section Biology of Archaea. Front Microbiol. 2020 Sep 18;11:584152, doi.org/10.3389/fmicb.2020.584152.


