The mechanism of degradation of mRNA
a) The mRNA decapping.
Decapping consists in the degradation of the cap structure protecting the 5’ end of mRNAs and is considered as the crucial and irreversible step before rapid degradation of mRNAs. Decapping is accomplished by the recruitment of a complex composed of the Dcp2 catalytic subunit and its activator Dcp1. This complex has a low intrinsic decapping activity and requires several accessory factors to be fully efficient. These include the Lsm1-Lsm7 complex that binds to the 3’ end of deadenylated mRNAs, also promotes decapping and Pat1 recruitment. Dhh1 and Edc1-3 proteins (Enhancer of Decapping) favour decapping. Finally, Dhh1, Scd6 and Pat1 have also been reported to mediate translational repression by limiting the formation of the 48S pre-initiation complex.
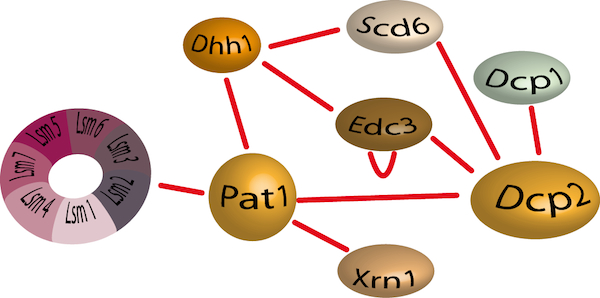
The decapping protein interaction network.
We have recently determined the crystal structure of the C-terminal domain from Pat1 (Fourati et al; 2014; Plos One), a scaffold protein responsible for the recruitment of either repressors of translation or enhancers of decapping activity, and identified the Edc3 region interacting with the Rps28 ribosomal protein (Kolesnikova et al; 2013; NAR). However, the mechanisms leading to the activation of decapping remain largely obscure. Interestingly, all these proteins are part of dynamic and multifunctional protein assemblies and the importance of these protein-protein interactions is becoming increasingly evident. It is therefore essential to understand in molecular details the dynamics of these complexes, and the interactions involved, in order to unravel the mechanisms of mRNA decay and the logic of its regulation.
Collaborator: Dr Bertrand Séraphin (IGBMC, CNRS, Univ. Strasbourg, Illkirch, France).
Our references related to this topics:
- Fourati Z; Kolesnikova O; Back R; Keller J; Charenton C; Taverniti V; Gaudon Plesse C; Lazar N; Durand D; van Tilbeurgh H; Séraphin B; Graille M. (2014) The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment. Plos One, 9(5), e96828.
- Kolesnikova O; Back R; Graille M; Séraphin B. (2013) Identification of the Rps28 binding motif from yeast Edc3 involved in the autoregulatory feedback loop controlling RPS28B mRNA decay. Nucleic Acids Res, 41(20), 9514-9523.
b) The Nonsense mediated mRNA decay or NMD pathway.
It is estimated that mRNAs containing premature stop codons (5-10% of eukaryotic mRNAs), are responsible for 1/3 of inherited genetic diseases such as Duchenne’s muscular dystrophy (DMD) or some cystic fibrosis, as well as various cancers. However, truncated proteins synthesized from these aberrant mRNAs are seldom detected due to the existence of a surveillance mechanism: the Nonsense mediated mRNA decay pathway (or NMD). The NMD pathway detects most of these aberrant mRNAs and destroys them rapidly from the pool of translatable mRNAs. Among the proteins implicated in this pathway are the release factors eRF1 and eRF3, the NMD-specific factors Upf1-3 and proteins involved in normal mRNAs.
We have determined the crystal structure of the N-terminal domain from Upf2 and identified a highly conserved region from this domain that is crucial for NMD, probably through the recruitment of a protein partner (Fourati et al; 2014; J. Mol. Biol / Fourati and Graille; 2014).
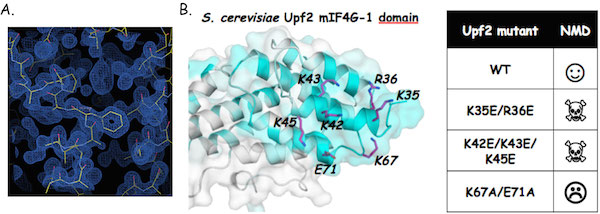
The Upf2 N-terminal domain is mandatory for NMD.
A. Experimental electron density maps obtained at 1.45Å resolution using the ARCIMBOLDO program (Courtesy from I. Uson). B. Highly conserved residues from Upf2 N-terminal domain are essential for NMD.
Collaborators: Pr Allan Jacobson (Univ. Massachusetts Medical School, Worcester, USA) & Pr Mans Ehrenberg (Uppsala Univ., Sweden)
Our references related to this topics:
- Fourati Z; Roy B; Millan C; Coureux PD; Kervestin S; van Tilbeurgh H; He F; Uson I; Jacobson A; Graille M. (2014) A highly conserved region essential for NMD in the Upf2 N-terminal domain. J Mol. Biol. 426(22), 3689-3702.
- Fourati Z & Graille M. (2014) Cytoplasmic mRNA surveillance pathways. In « Fungal RNA biology » (Springer International Publishing). Editors : A. Sesma and T. von der Haar. ISBN 978-3-319-05687-6.
c) Quality-control pathways degrading RNAs inducing strong translation stalls: the NGD, NSD and 18S-NRD pathways.
In addition to NMD, eukaryotic cells have evolved surveillance pathways aimed at degrading those RNAs that induce strong translational stalls during translation elongation. These are NGD (No-Go Decay), 18S-NRD (18S Nonfunctional rRNA decay) and NSD (Non-Stop Decay) pathways. They require the Dom34 and Hbs1 proteins, which are structurally similar to eRF1 and eRF3, respectively.
To understand the role of these two proteins in these surveillance mechanisms, we have solved the isolated structures of yeast Dom34 and Hbs1 proteins and obtained a low resolution model of the Dom34-Hbs1 complex using SAXS. In vivo experiments in yeast S. cerevisiae have shown that GTP nucleotide binding by Hbs1 is necessary for both NGD and 18S-NRD. Surprisingly, we have also demonstrated that the interaction between Dom34 and Hbs1 is required for NGD but not 18S-NRD (Graille et al; 2008; JBC / van den Elzen ; 2010; Nat. Struct. Mol. Biol. / Graille & Séraphin; 2012; Nat. Rev. Mol. Cell. Biol./ Fourati and Graille; 2014).
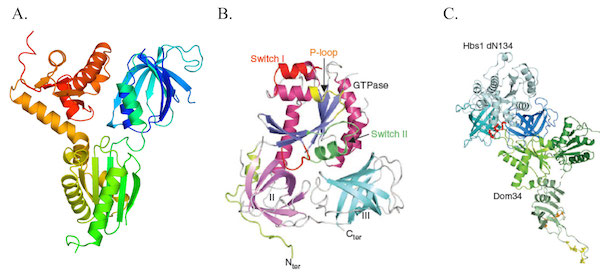
The Dom34-Hbs1 complex.
Representation of Dom34 (A; colored from blue (N-terminus) to red (C-terminus), Hbs1 (B) crystal structures and of the model of Hbs1-Dom34 obtained by SAXS (C).
Collaborator: Dr Bertrand Séraphin (IGBMC, CNRS, Univ. Strasbourg, Illkirch, France).
Our references related to this topics:
- Fourati Z & Graille M. (2014) Cytoplasmic mRNA surveillance pathways. In « Fungal RNA biology » (Springer International Publishing). Editors : A. Sesma and T. von der Haar. ISBN 978-3-319-05687-6.
- Graille M & Séraphin B. (2012) Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol., 13, 727-735 (Review).
- van den Elzen AMG, Henri J, Lazar N, Gas-Lopez M, Durand D, Lacroute F, NicaiseM, van Tilbeurgh H, Séraphin B, Graille M. (2010) Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat. Struct. Mol. Biol. 17(12), 1446-53.
- Graille M, Chaillet M, van Tilbeurgh H. (2008) Structure of yeast Dom34: a protein related to translation termination factor eRF1 and involved in No-Go decay. J Biol Chem. 283(11), 7145-54.


