Post-transcriptional and post-translational modifications of factors (RNA or proteins) involved in translation.
a) Trm112, a central player involved in maturation of various factors involved in translation.
Trm112, a small zinc finger protein of 15 kDa, is known to interact with, and to activate, four methyltransferases (MTases), namely: Mtq2, Trm9, Trm11 and Bud23, all related to ribosome function.
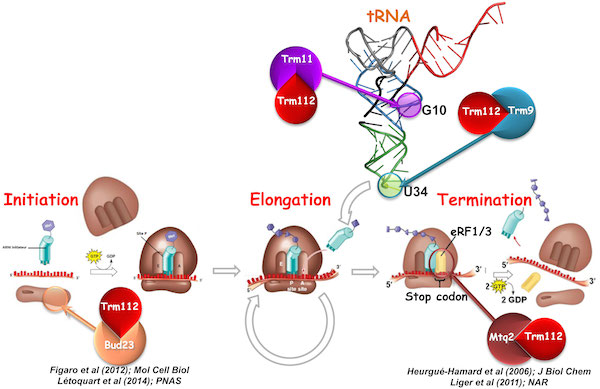
The Mtq2-Trm112 complex methylates the translation termination factor eRF1 on the glutamine side chain of its universally conserved GGQ motif, which is directly involved in the release of newly synthesized proteins (Heurgué-Hamard et al; 2006; JBC / Liger et al; 2011; NAR). The Bud23-Trm112 complex is implicated in ribosome biogenesis by methylating guanosine 1575 of 18S rRNA (Figaro et al; 2012; Mol Cell Biol / Létoquart et al; 2014; PNAS). Trm11-Trm112 and Trm9-Trm112 complexes modify tRNA (Létoquart et al; 2015; NAR). Trm112 is then truly unique acting as an activating platform of four MTases involved in rRNA, tRNA and translation factor modifications, ideally placing it at the interface between ribosome synthesis and function. In human, some of these proteins have been involved in diseases such as cancers.
Our goals are to characterize the mechanism of substrate recognition and modification by these Trm112-MTase complexes in order to enlighten the biological impact of these modifications.

Collaborators: Dr Valérie Heurgué-Hamard (IBPC, CNRS, Paris, France) & Pr Denis L.J. Lafontaine (Université Libre de Bruxelles, Charleroi, Belgium).
Our references related to this topics:
- Létoquart J, Tran N.V, Caroline V, Alexandrov A, Lazar N, van Tilbeurgh H, Liger D#, Graille M#. (2015) Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res, in press.
- Létoquart J; Huvelle E; Wacheul L; Bourgeois G; Zorbas C; Graille M; Heurgué-Hamard V; Lafontaine D.L.J. (2014) Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci U S A. 111(51), E5518-26.
- Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DLJ, Heurgué-Hamard. (2012) Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell. Biol. 32(12), 2254-67.
- Graille M, Figaro S, Kervestin, S, Buckingham RH, Liger D, Heurgué-Hamard V. (2012) Methylation of class I translation termination factors : structural and functional aspects. Biochimie. 94, 1533-43 (Review).
- Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgué-Hamard V, Graille M. (2011) Mechanism of activation of methyltransferases involved in translation by the Trm112 « hub » protein. Nucleic Acids Res. 39(14), 6249-59.
- Heurgué-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. (2006) The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem. 281(47), 36140-8.
- Graille M, Heurgue-Hamard V, Champ S, Mora L, Scrima N, Ulryck N, van Tilbeurgh H, Buckingham RH. (2005) Molecular Basis for Bacterial Class I Release Factor Methylation by PrmC. Mol. Cell. 20(6), 917-27.
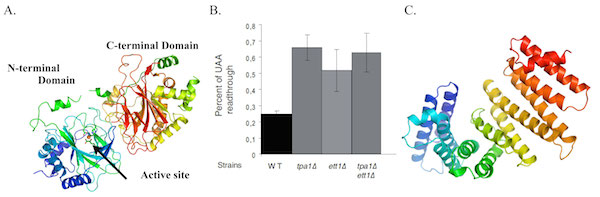
b) The prolyl-hydroxylase Tpa1 and its partner Ett1 regulate translation termination efficiency
The Tpa1 protein has been characterized in S. cerevisiae yeast as a component (together with eRF1, eRF3 and Pab1) of a mRNP bound to the mRNA 3’ end and to influence stop codon recognition.
To investigate its function, we have solved the crystal structure of S. cerevisiae Tpa1, revealing a strong structural similarity with prolyl-4-hydroxylases (enzymes catalyzing hydroxylation of proline side chains in an O2-dependent manner) and identified Ett1 as a new factor influencing stop codon read-through (Henri et al; 2010; JBC). It was later shown that Tpa1 is indeed a prolyl hydroxylase modifying a ribosomal protein on a conserved proline residue. In addition, we have solved the structure of Nro1, the Ett1 orthologue from S. pombe, and shown that this protein acts in the same pathway as Tpa1 and influences the accuracy of stop codon recognition (Rispal et al; 2011; RNA).
Collaborator: Dr Bertrand Séraphin (IGBMC, CNRS, Univ. Strasbourg, Illkirch, France).
Our references related to this topics:
- Henri J, Rispal D, Bayart E, van Tilbeurgh H, Séraphin B, Graille M. (2010) Structural and functional insights into S. cerevisiae Tpa1, a putative prolyl hydroxylase influencing translation termination and transcription. J Biol Chem, 285(40), 30767-78.
- Rispal D, Henri J, van Tilbeurgh H, Graille M, Séraphin B. (2011) Structural and functional analysis of Nro1/Ett1 : a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe. RNA. 17(7), 1213-24.


