Mécanismes moléculaires de l'adaptation microbienne
Our web site has moved to this address: https://lob.ip-paris.fr
Researchers
Alexey ALEKSANDROV, CR, CNRS
Hubert BECKER, PU, Sorbonne université
Lionel GUITTAT, MdC, Université Sorbonne Paris-Nord
Ursula LIEBL, DR, CNRS
Roxane LESTINI, Prof, Ecole Polytechnique
Jean-Louis MERGNY, DR, Inserm
Hannu MYLLYKALLIO, DR, CNRS
Stephane SKOULOUBRIS, MdC, Univ. Paris–Saclay
Technicians & engineers
Zackie AKTARY
Anne CUCCHIARINI
Isabelle LAMARRE-JOUENNE
Caroline L'HERMITTE STEAD
Lucille MELLOTTEE
Thesis and Master students
Anaïs BAYARD
Tom GARNIER (Master 2)
Ahlam HAKAM (Master 2)
Maria IMEZAR
Yu LUO
Dorian NOURY
Jiawei WANG
Rongxin ZHANG
For inquiries on the different research projects and possibilities for joining us, please contact us by email (Nous contacter)
Research topics
Our principal research goals are to understand how microorganisms adapt to different metabolic changes, crucial for their survival. Our studies concern sensing and signaling of different gas conditions, the availability of DNA building blocks, and the reciprocal interactions of DNA replication and repair, responsible for guaranteeing genomic integrity at extreme conditions. We are also interested in the implication of the unusual DNA/RNA structures in key biological processes. To address these crucial questions, we use a wide range of experimental and computational methods.
Ongoing projects
Our research interests include five major topics:
Much recent work has been devoted to molecular biological and biochemical studies to unravel the reaction mechanism and inhibition of alternative thymidylate synthase ThyX, discovered in the laboratory. ThyX proteins are essential for de novo synthesis of the letter “T” (thymidylate) in DNA in many prokaryotes. Our work has shed new light on the evolution of prokaryotic genomes, since the low catalytic activity of ThyX limits DNA replication and consequently is found in prokaryotic species with small genomes, whereas organisms with more complex genomes use thymidylate synthase ThyA, the enzyme present also in humans. As ThyX is present in many human pathogens such as Helicobacter pylori or Mycobacterium tuberculosis but absent in human cells, this opens the way to the development of novel antimicrobial strategies by identifying new inhibitors of this enzyme. In collaboration with Team 2 (LOB), several complementary techniques have been applied to research on ThyX. In particular, we have observed unusually high flexibility of the active site of ThyX. We are currently interested in evolutionary histories of the thymidylate synthase families. We have proposed how complex substrate interactions modulate oxygen reactivity of ThyX proteins. We are also analyzing folate metabolic and related pathways in the human pathogenic bacterium H. pylori. Finally, we have obtained proof–of-concept using animal models of infection for the anti-microbial activity of 1,4-naphthoquinones that we patented as efficient and specific inhibitors of the alternative thymidylate synthase X (Figure 1). Our primary goal is to render our inhibitor series for eventual preclinical development through collaborative efforts. This will require synthesis of new lead derivatives to optimize anti-microbial activity of already identified compounds.
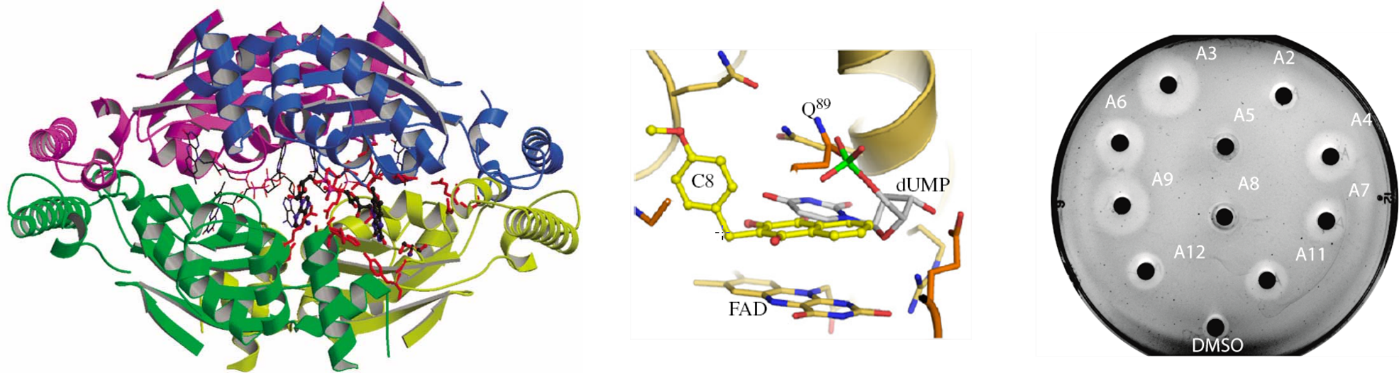
Figure 1: 3D structure of ThyX (left); Representation of the C8-C1 inhibitor fitted into ThyX catalytic site (middle) (adapted from Basta et al (2012); Antibiotic susceptibility testing of ThyX inhibitors (right).
-
Modranka J, Li J, Parchina A, Vanmeert M, Dumbre S, Salman M, Myllykallio H, Becker HF, Vanhoutte R, Margamuljana L, Nguyen H, Abu El Asrar R, Rozenski J,Herdewijn P, Dejonghe S, Lescrinier E. Synthesis and structure-activity relationship studies of benzo[b][1,4]oxazin-3(4H)-one analogues as inhibitors of mycobacterial thymidylate synthase X. ChemMedChem. 2019 Jan 31. doi: 10.1002/cmdc.201800739. [Epub ahead of print]
-
Sodolescu A, Dian C, Terradot L, Bouzhir-Sima L, Lestini R, Myllykallio H, Skouloubris S, Liebl U. Structural and functional insight into serine hydroxymethyltransferase from Helicobacter pylori. PLoS One. 2018 Dec 14;13(12):e0208850. doi: 10.1371/journal.pone.0208850.
-
Aleksandrov A, Myllykallio H. Advances and challenges in drug design against tuberculosis: application of in silico approaches. Expert Opin Drug Discov. 2019 Jan;14(1):35-46. doi: 10.1080/17460441.2019.1550482.
-
Myllykallio H, Sournia P, Heliou A, Liebl U (2018). Unique Features and Anti-microbial Targeting of Folate- and Flavin-Dependent Methyltransferases Required for Accurate Maintenance of Genetic Information. Front Microbiol. 9:918. doi: 10.3389/fmicb.2018.00918 (review)
-
Abu El Asrar R, Margamuljana L, Klaassen H, Nijs M, Marchand A, Chaltin P, Myllykallio H, Becker HF, De Jonghe S, Herdewijn P, Lescrinier E (2017). Discovery of a new Mycobacterium tuberculosis thymidylate synthase X inhibitor with a unique inhibition profile Biochem Pharmacol. 135:69-78. doi: 10.1016/j.bcp.2017.03.017.
-
Djaout K, Singh V, Boum Y, Katawera V, Becker HF, Bush NG, Hearnshaw SJ, Pritchard JE, Bourbon P, Madrid PB, Maxwell A, Mizrahi V, Myllykallio H, Ekins S (2016). Predictive modeling targets thymidylate synthase ThyX in Mycobacterium tuberculosis. Sci Rep. 6:27792. doi: 10.1038/srep27792. PubMed PMID: 27283217; PubMed Central PMCID: PMC4901301.
-
Garg D, Skouloubris S, Briffotaux J, Myllykallio H, Wade RC (2015). Conservation and Role of Electrostatics in Thymidylate Synthase. Sci Rep. 5:17356.doi: 10.1038/srep17356. PubMed PMID: 26612036; PubMed Central PMCID: PMC4661567.
-
Skouloubris S, Djaout K, Lamarre I, Lambry JC, Anger K, Briffotaux J, Liebl U, de Reuse H, Myllykallio H (2015). Targeting of Helicobacter pylori thymidylate synthase ThyX by non-mitotoxic hydroxy-naphthoquinones. Open Biol. 6:150015. doi: 10.1098/rsob.150015.
-
Salo-Ahen OM, Tochowicz A, Pozzi C, Cardinale D, Ferrari S, Boum Y, Mangani S, Stroud RM, Saxena P, Myllykallio H, Costi MP, Ponterini G, Wade RC (2015). Hotspots in an obligate homodimeric anticancer target. Structural and functional effects of interfacial mutations in human thymidylate synthase. J Med Chem. 58(8):3572-81.
-
International patents PCT N°PCT/EP201110555 and BET10P0152 on antimicrobial activity of ThyX inhibitors (INSERM/CNRS)
How Archaea, the third domain of Life, replicate their DNA and maintain their genomes intact often under deleterious environmental conditions is also investigated by our team.
A. Replication Dynamics: We have established the salt-loving archaea Haloferax volcanii as a unique model for studying archaeal DNA replication and investigate the function of the helicase/nuclease Hef in replication restart. By expressing replication proteins fused to the Green Fluorescent protein, and using fluorescence microscopy technics, we study their localization in living cells to better understand replication dynamics in these highly polyploidy archaeal cells (Figure 2A).
B. Circular RNAs: A project on understanding the widespread functional roles of circular RNAs in hyperthermophilic archaea is underway. Circular RNAs, which have recently emerged, are formed by ligation of the free ends of linear RNA molecules by an atypic homodimeric RNA ligase (Rnl3 family). Physiological circular RNAs were identified, in P. abyssi cells, by RNAseq experiments, and we investigated the physiological significance of this unexpected RNA circularization activity (Figure 2B).
C. Characterization of a non-canonical Mismatch Repair System: study of the NucS (EndoMS) protein key enzyme. The NucS protein discovered and characterized for the first time in the laboratory exhibits nuclease activity on single-stranded and branched DNA but also has the striking feature of recognizing and cleaving mismatched DNAs. We have thus shown that the deletion of the nucS gene in the bacterium of industrial interest Corynebacterium glutamicum, increased the frequency of spontaneous mutation occurrence by at least a 200-fold. The NucS protein is found both in some archaeal species (eg. Pyrococcus furiosis, Thermococcus kodakarensis, Haloferax volcanii) but also in the Actinonacteria phylum (Mycobacterium sp., Corynebacterium sp., Streptomyces sp.) Experiments are currently being conducted to determine the precise role of this protein in vivo and to identify protein partners also involved in this non-canonical mismatch repair system (Figure 2C).
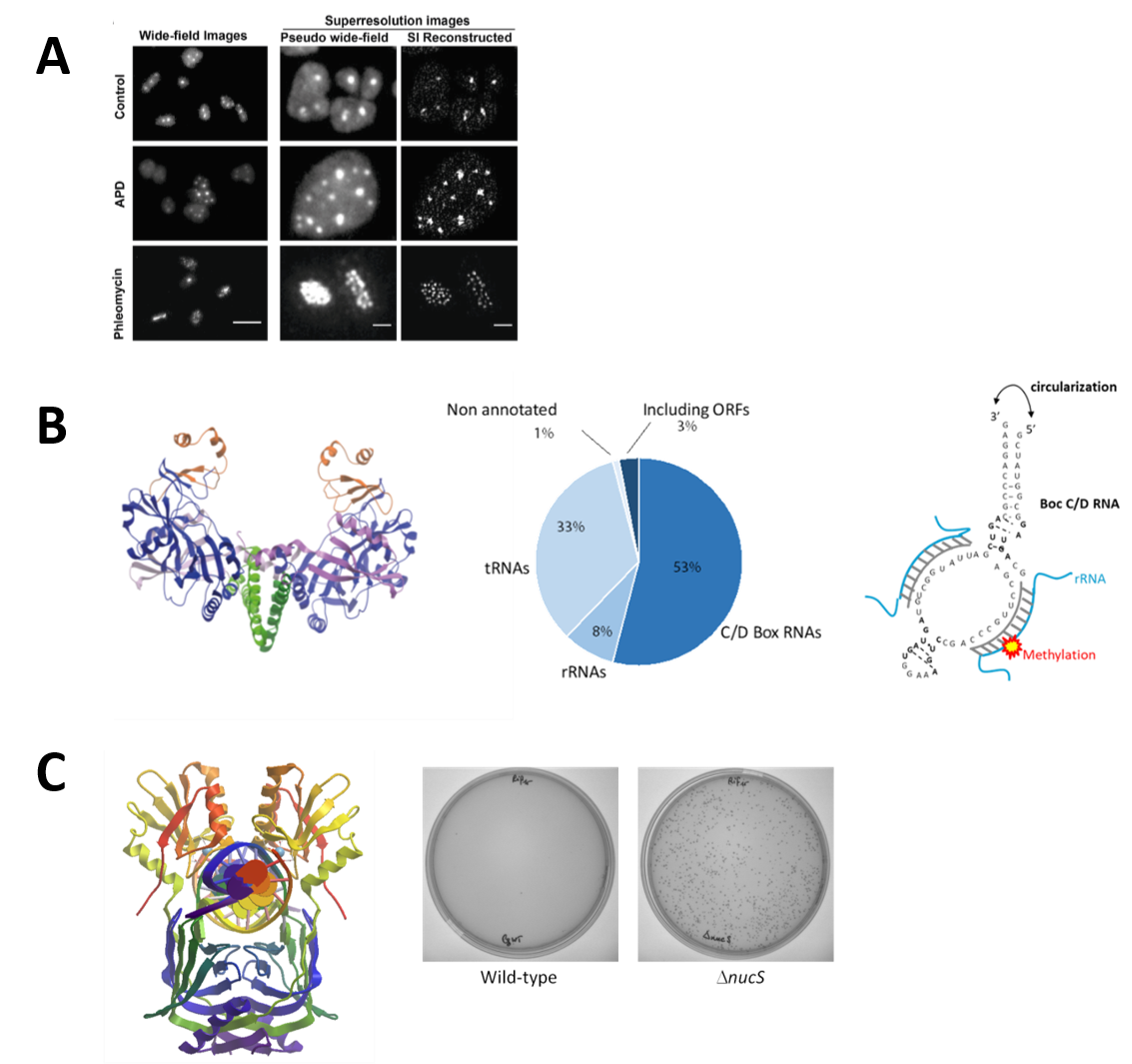
Figure 2:
-
The quantification and localization of archaeal replication factories in live Haloferax volcanii cells using super-resolution microscopy (adapted from Delpech et al., 2018)
-
Structure of RNA ligase Pab1020 (PDB code 2VUG) (left); Representation of the different functional circular RNA classes obtained for Pyrococcus abyssi (RNAseq) (middle); Schematic of the structure of an archaeal Box C/D RNA (right) (adapted from Becker et al., 2017 )
-
3D NucS structure (dimer) of Thermococcus kodakarensis co-crystallized with DNA having a G:T mismatch (PDB:5GKE) (left); Increased frequency of occurrence of spontaneous rifampin-resistant mutants in a nucS-deficient strain (Dnucs) of Corynebacterium glutamicum (right) (adapted from Ishino et al (2018)).
Very recently, we have been involved in designing and developing methods of multiplex genome editing in collaboration with the Church laboratory at U. Harvard (thesis of O. Castanon, co-supervision, Smith et al., 2020).
-
Lestini R, Collien Y, Olivier D, Olivier N, Myllykallio, H. BrdU Incorporation and Labeling of Nascent DNA to Investigate Archaeal Replication Using Super-Resolution Imaging. Methods Mol Biol . 2022 2522:419-434. doi: 10.1007/978-1-0716-2445-6_29.
-
Cockram C, Thierry A, Gorlas A, Lestini R, Koszul R. Euryarchaeal genomes are folded into SMC-dependent loops and domains, but lack transcription-mediated compartmentalization. Mol Cell. 2021 Feb 4;81(3):459-472.e10. doi: 10.1016/j.molcel.2020.12.013. Epub 2020 Dec 30. PMID: 33382984.
-
Smith CJ, Castanon O, Said K, Volf V, Khoshakhlagh P, Hornick A, Ferreira R, Wu CT, Güell M, Garg S, Ng AHM, Myllykallio H, Church GM. Enabling large-scale genome editing at repetitive elements by reducing DNA nicking. Nucleic Acids Res. 2020 May 21;48(9):5183-5195. doi: 10.1093/nar/gkaa239.
-
Castanon O, Cory JS, Khoshakhlagh P, Ferreira R, Güell M, Said K, Yildiz R, Dysart M, Wang S, Thompson D, Myllykallio H, George M. Church CRISPR-mediated biocontainment doi: https://doi.org/10.1101/2020.02.03.922146 (preprint).
-
Becker HF, L'Hermitte-Stead C, Myllykallio H. Diversity of circular RNAs and RNA ligases in archaeal cells. Biochimie. 2019 Sep;164:37-44. doi: 10.1016/j.biochi.2019.06.011.
-
Smith CJ, Castanon O, Said K, Khoshakhlagh P, Volf V, Hornick A, Ferreira R, Wu CT, Güell M, Myllykallio H, Church GM (2019) Enabling large-scale genome editing by reducing DNA nicking. Biorxiv (preprint).
-
Delpech, F., Collien, Y., Mahou, P., Beurepaire, E., Myllykallio, H., Lestini, R. (2018) Snapshots of Archaeal DNA Replication and Repair in Living Cells Using Super-resolution Imaging. Nucleic Acid Res. 16:10757-10770. doi: 10.1093/nar/gky829.
-
Ishino S, Skouloubris S, Kudo H, l'Hermitte-Stead C, Es-Sadik A, Lambry JC, Ishino Y, Myllykallio H (2018). Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res. 46(12):6206-6217.
-
Khallef S, Lestini R, Myllykallio H, Houali K (2018). Isolation and identification of two extremely halophilic archaea from sebkhas in the Algerian Sahara. Cell Mol Biol. 31;64(4):83-91.
-
Becker HF, Héliou A, Djaout K, Lestini R, Regnier M, Myllykallio H (2017). High-throughput sequencing reveals circular substrates for an archaeal RNA ligase. RNA Biol. 1-11. doi: 10.1080/15476286.2017.1302640.
-
Lestini R, Delpech F, Myllykallio H. DNA replication restart and cellular dynamics of Hef helicase/nuclease protein in Haloferax volcanii (2015). Biochimie. pii: S0300-9084(15)00230-8 (review).
Bacterial heme sensor and flavin-containing proteins
Our third research avenue concerns bacterial adaptation to different gas concentrations and the implication of heme-based gas sensor proteins. These proteins are essential components of signal transduction chains and the binding of gas molecules induces structural changes that lead to adaptation of the activity of other proteins and specific changes in gene expression. We aim at understanding the mechanism of signaling initiation and propagation, as well as the dynamic events that lead to protein function, in particular of carbon monoxide (CO) sensors.
A second activity concerns flavin-dependent methylation/demethylation enzymes of nucleic acids, in particular ThyX, TrmFO and LSD1. ThyX and TrmFO use an FAD cofactor as covalent catalyser; a strategy that appears to be more widespread than initially thought and which we are exploring. The human flavoenzyme lysine-specific histone demethylase LSD1 catalyses the specific demethylation of lysines 4 and 9 of histone H3 and thus creates MTHF from its substrate H4folate: the inverse reaction of thymidylate synthase ThyX. In collaboration with M. Vos (LOB, team 2) we study the dynamics of internal electron transfer and try to get detailed insight into the LSD1 reaction mechanism.
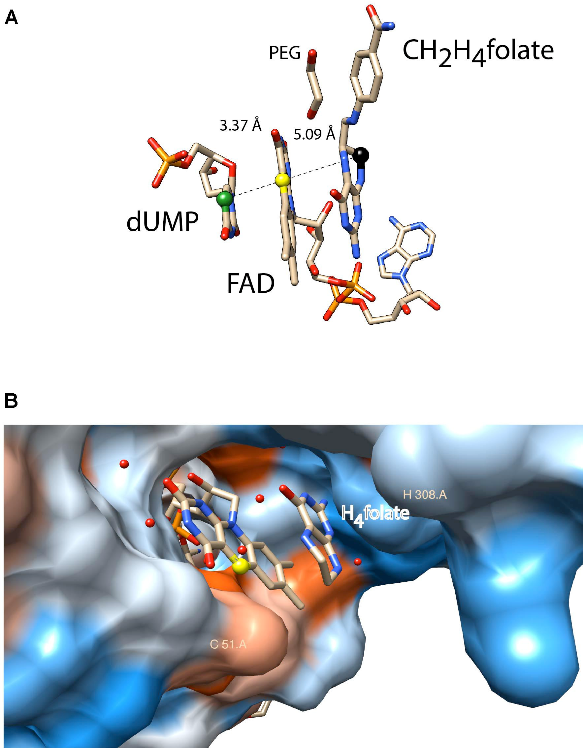
Figure 3: Active site of ThyX and TrmFO proteins. (A) Binding configuration and stacking interactions of dUMP, FAD, and CH2H4folate in the active site of Thermotoga maritima ThyX (PDB: 4GT9). The proposed one-carbon relay from CH2H4folate (black ball) via the N5 of the FAD (yellow ball) to the accepting C5 of the uracil ring is depicted. (B) Active site of monomeric Thermus thermophilus TrmFO with a fragment of tetrahydrofolate (PDB: 3G5R).
-
Interaction of the Full-Length Heme-Based CO Sensor Protein RcoM-2 with Ligands. Salman M, Villamil Franco C, Ramodiharilafy R, Liebl U, Vos MH. Biochemistry. 2019 Oct 1;58(39):4028-4034. doi: 10.1021/acs.biochem.9b00623. PMID: 31502443
-
Structural and functional insight into serine hydroxymethyltransferase from Helicobacter pylori. Sodolescu A, Dian C, Terradot L, Bouzhir-Sima L, Lestini R, Myllykallio H, Skouloubris S, Liebl U. PLoS One. 2018 Dec 14;13(12):e0208850. doi: 10.1371/journal.pone.0208850. eCollection 2018. PMID: 30550583
-
Ultrafast photochemistry of the bc1 complex. Vos MH, Reeder BJ, Daldal F, Liebl U. Phys Chem Chem Phys. 2017 Mar 1;19(9):6807-6813. doi: 10.1039/c7cp00193b. PMID: 28218331
-
Identification of the TyrOH•+ Radical Cation in the Flavoenzyme TrmFO. Nag L, Sournia P, Myllykallio H, Liebl U, Vos MH. J Am Chem Soc. 2017 Aug 23;139(33):11500-11505. doi: 10.1021/jacs.7b04586. PMID: 28745052
-
Thermal stability and binding energetics of thymidylate synthase ThyX. Krumova S, Todinova S, Tileva M, Bouzhir-Sima L, Vos MH, Liebl U, Taneva SG. Int J Biol Macromol. 2016 Oct;91:560-7. doi: 10.1016/j.ijbiomac.2016.05.083. PMID: 27268384
-
Ultrafast Spectroscopy Evidence for Picosecond Ligand Exchange at the Binding Site of aHeme Protein: Heme-Based Sensor YddV. Lambry JC, Stranava M, Lobato L, Martinkova M, Shimizu T, Liebl U, Vos MH. J Phys Chem Lett. 2016 Jan 7;7(1):69-74. doi: 10.1021/acs.jpclett.5b02517. PMID: 26651267
-
Unusual Dynamics of Ligand Binding to the Heme Domain of the Bacterial CO Sensor Protein RcoM-2. Bouzhir-Sima L, Motterlini R, Gross J, Vos MH, Liebl U. J Phys Chem B. 2016 Oct 20;120(41):10686-10694. doi: 10.1021/acs.jpcb.6b08160. PMID: 27661620
-
H. Open Biol. 2015 Jun;5(6):150015. doi: 10.1098/rsob.150015. PMID: 26040760
-
Kinetic Analysis of a Globin-Coupled Histidine Kinase, AfGcHK: Effects of the Heme Iron Complex, Response Regulator, and Metal Cations on Autophosphorylation Activity. Fojtikova V, Stranava M, Vos MH, Liebl U, Hranicek J, Kitanishi K, Shimizu T, Martinkova M. Biochemistry. 2015 Aug 18;54(32):5017-29. doi: 10.1021/acs.biochem.5b00517. PMID: 26212354
-
Time-resolved infrared spectroscopic studies of ligand dynamics in the active site from cytochrome c oxidase. Vos MH, Liebl U. Biochim Biophys Acta, 2015 Jan;1847(1):79-85. doi: 10.1016/j.bbabio.2014.07.018.
Quadruplexes are unusual nucleic acid structures formed by G-rich DNA and RNA sequences and based on the stacking of two of more G-quartets (Figure 4). G-quadruplexes can be stable under physiological conditions and the evidence for quadruplex formation in vivo is now compelling. Using a prediction algorithm (G4Hunter) developped in house, we have analyzed quadruplex propensity in a variety of genomes including viruses, eukaryotic pathogens and Archaea. We are especially interested in the implication of these structures in key biological processes and how quadruplexes may be recognized by specific small ligands or proteins.
Figure 4:A G-quartet (top) and G-quadruplexes of various strand stoichiometries (bottom).
-
Gazanion E, Lacroix L, Alberti P, Gurung P, Wein S, Cheng M, Mergny JL, Gomes AR, Lopez-Rubio JJ. Genome wide distribution of G-quadruplexes and their impact on gene expression in malaria parasites. PLoS Genet. 2020, 16 (7) e1008917.
-
Brázda V, Luo Y, Bartas M, Kaura P, Porubiaková O, Šťastný J, Pečinka P, Verga D, Da Cunha V, Takahashi TS, Forterre P, Myllykallio H, Fojta M, Mergny JL. G-quadruplexes in the Archaea domain. Biomolecules. 2020, 10: 1349. doi:10.3390/biom10091349
-
Carvalho J, Lopes-Nunes J, Campello MPC, Paulo A, Milici J, Meyers C, Mergny JL, Salgado GF, Queiroz JA, Cruz C. Human papillomavirus G-rich regions as potential antiviral drug targets. Nucleic Acids Therapeutics. 2021; in press. doi: 10.1089/nat.2020.0869
-
Mergny JL*, Guittat L, Ségal-Bendirdjian E. Télomères et Télomérase : des cibles toujours pertinentes en oncologie ? Bull Cancer. 2021, 108 :30-54.
-
Busto N, García-Calvo J, Vicente Cuevas J, Herrera A, Mergny JL, Pons S, Torroba T, García B. Influence of core extension and side chain nature in targeting G-quadruplex structures with perylene monoimide derivatives. Bioorg Chem. 2021, in press. https://doi.org/10.1016/j.bioorg.2021.104660
-
Miron CE, van Staalduinen L, Rangaswamy AM, Chen M, Liang Y, Jia Z, Mergny JL*, Petitjean A. Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations. Angew Chem Int Ed Engl. 2021, 60, 2500-2507.
-
Stadlbauer P, Islam B, Otyepka M, Chen J, Monchaud D, Zhou J, Mergny JL, Sponer J. Insights into G-Quadruplex–Hemin Dynamics Using Atomistic Simulations: Implications for Reactivity and Folding. J Chem Theory Comput 2021, in press: https://dx.doi.org/10.1021/acs.jctc.0c01176
-
Carson JJK, Miron CE, Luo J, Mergny JL, van Staalduinen L, Jia Z, Petitjean A. Exploration of head-to-tail and head-to-head isomers of a guanine quadruplex platinum-based binder. Inorg. Chim. Acta. 2021, 518, 120236 https://doi.org/10.1016/j.ica.2020.120236
-
Chen J, Cheng M, Wang J, Qiu D, Monchaud D, Mergny JL, Ju H, Zhou J. The catalytic properties of G-quadruplexes rely on their structural integrity. Chinese J Catalysis. 2021, 42, 1102-1107. doi: 10.1016/S1872-2067(20)63744-5.
-
Aznauryan M, Noer SL, Pedersen CW, Mergny JL, Teulade-Fichou MP, Birkedal V. Ligand binding to dynamically populated G-quadruplex DNA. ChemBioChem. 2021, in press. doi: 10.1002/cbic.202000792.
-
Luo Y, Granzhan A, Verga D & Mergny JL. FRET-MC : a fluorescence melting competition assay for studying G4 structures in vitro. Biopolymers. 2021, in press https://doi.org/10.1002/bip.23415
-
Figueiredo J, Miranda A, Lopes-Nunes J, Carvalho J, Alexandre D, Valente S, Mergny JL, Cruz C. Targeting nucleolin by G-quadruplex forming motif. Biochem Pharmacol. 2021, in press. doi: 10.1016/j.bcp.2021.114418
Computational chemistry
This direction of the research is tightly connected with the other on-going research projects. In particular, using methods of computational structural bioinformatics in synergy with spectroscopic experiments, we are investigating the mechanism of inhibition of bacterial ThyX by the inhibitors, such as 1,4- naphthoquinones discovered in our team in order to propose better inhibitors with the ultimate goal to design better antibiotics. Using this combined approach, we recently discovered benzoate as a weak inhibitor of ThyX from Mtb, shown in Figure 5, which can serve as a scaffold for the future rational design endeavors.
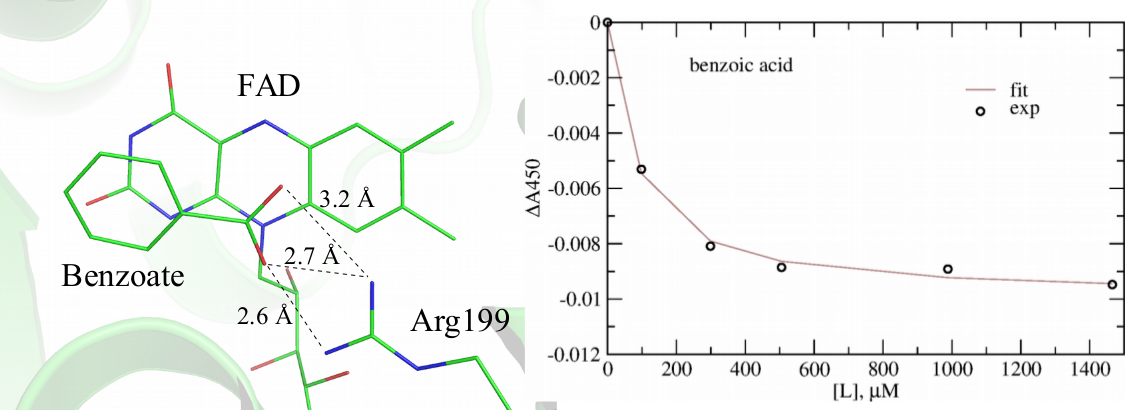
Figure 5. Identification benzoate as a week inhibitor of ThyX from Mtb. Left: modeled structure of benzoate in complex with ThyX; Right: spectrophotometric titration of ThyX with benzoate demonstrating its protein binding.
Recent publications
-
Aleksandrov A, Roux B., and MacKerell, Jr. AD, pKa Calculations with the Polarizable Drude Force Field and Poisson-Boltzmann Solvation Model, J. Chem. Theory Comput. 2020, 16, 7, 4655–4668
-
Aleksandrov A. A Molecular Mechanics Model for Flavins. J Comput Chem. 2019; 40(32):2834-2842.
-
Aleksandrov A, Myllykallio H. Advances and challenges in drug design against tuberculosis: application of in silico approaches. Expert Opinion on Drug Discovery, 2018, pp.1 - 12.
-
Aleksandrov A, Lin FY, Roux B, MacKerell AD Jr. Combining the polarizable Drude force field with a continuum electrostatic Poisson-Boltzmann implicit solvation model. J Comput Chem. 2018; 39(22):1707-1719.
Technical approaches
In silico methods and phylogenomics, microbiology and cellular biology, molecular biology, protein biochemistry and enzymology, inhibitory screening, rapid mixing spectroscopic techniques; steady-state and time-resolved spectroscopy (absorption, fluorescence including FRET), life cell imaging, high throughput sequencing (DNA, RNA), bioinformatics (quadruplex predictions), computational structural bioinformatics.
Collaborations 2010-2020:
A. Maxwell, John Innes Centre, UK (publication 2016)
B. Golinelli, Collège de France, Paris (ANR PCV publications 2010, 2011)
C. Cruz, Univ. Beira Interior, Covilha, Portugal (8 publications since 2017)
D. Flament and G. Henneke, IFREMER (ANR Blanc, publications 2007-2012)
D. Lechardeur, A. Gruss, INRA UMR 1319, Jouy-en-Josas (publication 2012)
E. Lescrinier, U. Leuven, Belgium (1 publication 2017, 2019)
G.M. Church, Harvard Medical School (thesis in co-supervision, NAR 2020, 2 preprints)
G. Marverti, U. Modena, Italie (publications 2012, 2014)
H. de Reuse, I. Pasteur (ANR Emergence, publication 2015)
H. van Tilbeurgh and M. Graille, U. Paris Sud, Orsay (ANR EMERGENGE, publications 2006, 2008, 2012)
J. Zhou, Nanjing University, China (14 publications since 2012)
J. Eaton-Rye, H. Luo, U. Otago, New Zealand (F-NZ Lottery Health Grant; publication 2011)
J. Guillon, Université de Bordeaux (6 publications 2017-2020)
J. Sponer, Institute of Biophysics, Brno, Czech Republic (3 publications since 2018)
L. Lacroix, ENS, Paris (8 publications since 2011)
L. Trantirek, Institute of Biophysics, Brno, Czech Republic (3 publications since 2018)
M. Lavigne, Institut Pasteur, Paris (2 publications submitted in 2020)
M-P. Teulade-Fichou, Institut Curie, Orsay (14 publications since 2011)
M.A. Hink, EuroBioimaging/U. Amsterdam, The Netherlands (publication 2013)
P. R. Ortiz de Montellano, A. Ioanoviciu, UCSF, USA (publication 2011)
R. Wade, Heidelberg Institute for Theoretical Studies and U. Heidelberg, Germany (review 2010, publication 2015)
V. Brazda, Institute of Biophysics, Brno, Czech Republic (8 publications since 2017)
S. Krumova, Bulgarian Academy of Sciences, IBBME, Sofia, Bulgaria (RILA 2013-2014, coordinator U. Liebl)
Y. Ishino, Kyushu University, Japan (publications 2001, 2018)
Former members including current affiliation
Véronique BALLAND (post-doctoral fellow, 2003-2004), now MdC Paris VII
Tamara BASTA (post-doctoral fellow 2008-2010), now MdC Paris XI
Yap BOUM (PhD student 2006-2008), now research director, Epicentre, Uganda
Julien BRIFFOTAUX (post-doctoral fellow 2011-1012)
Yoann COLLIEN (PhD student 2016-2019), now High school Biology teacher
Oscar CASTANON (PhD student 2016-2019)
Floriane DELPECH (PhD student 2013-2016)
Frédéric ESCARTIN (PhD student 2004-2008), post-doc INRA
Sébastien GRAZIANI (PhD student 2002-2005), now researcher at DGA
Klara HOLA (Marie Curie fellow, 2004)
Magda KOSMOPOULOU (Marie Curie fellow, 2006)
Damien LEDUC (PhD student 2001-2004), Professor of Biology, Agrégé en Biochimie Génie Biologique
Alessio LIGABUE (visiting student, post-doctoral fellow 2008-2010), now research associate at U. Washinton, Seattle
Gérard LIPOWSKI (PhD student 1999-2003), now managing director at Quintesensia, Tokyo, Japan
Laurence MESLET-CLADIERE (post-doctoral fellow, 2003-2006), now MdC U. Bretagne
Cédric NORAIS (PhD student 2003-2007), now MdC E. polytechnique
Mayla SALMAN (PhD student 2016-2019)
Andreea SODOLESCU (TODEA) (PhD student 2006-2009), now Medical Science Liaison Officer, Chugai Pharma France
John ULMER (post-doctoral fellow, 2007-2009), post-doc Paris VII.
